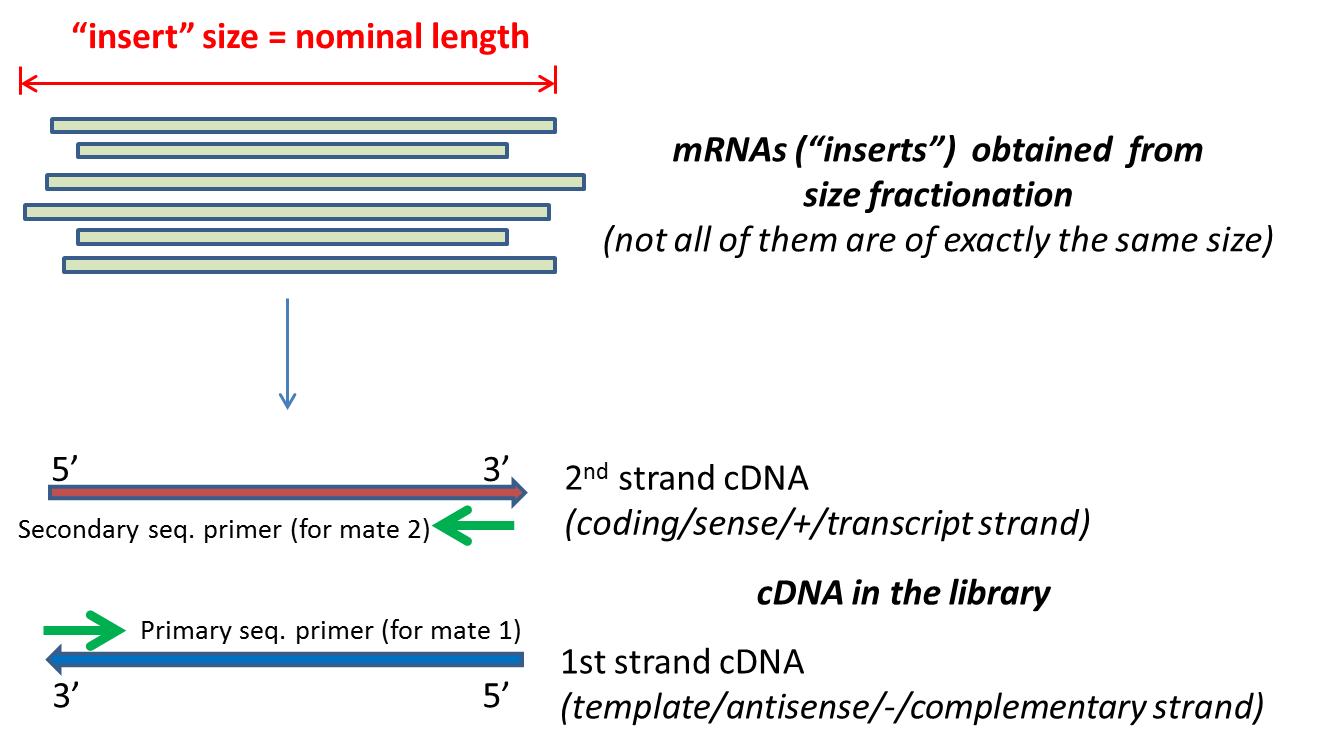
paired end sequencing insert size
Using the latest Illumina platform the MiSeq paired end reads of. Nine percent of read pairs were found to have an insert size of less than 250 bp and so likely to contain adapter after mapping to the reference genome of which 98 were also found to have an insert size of less than 250 bp by overlapping the reversed sequence.
What Is Paired End 150 Omega Bioservices
So why you get reads of the same length when you sequence.
. Including flanking intronic regions of diagnostic significance eg due to splice variants of about 30bp at either side the average target of interest is about 220 bp long. These reads can then be sequenced using the same SP1-SP2 adapter protocols used in PE sequencing. The reads have a length of typically 50 - 300 bp.
Multiple insert size paired-end sequencing for deconvolution of complex transcriptomes Authors. The difference in insert size stems from the difference in protocols. The fragment size which you need to select for during a gel purification for example would be the insert size length of both adapters around 120 bp extra for both Illumina.
Paired-end sequencing also provides greater ability to overcome the obstacles of characterizing repetitive sequence ele-ments by filling in gaps of consensus sequence to achieve complete. Analyze Store and Share in Illuminas BaseSpace Cloud Data analysis is an important factor to consider for sequencing applications. To read more about the different parts of a prepared DNA fragment please take a look on the figure in the article about observed sequence lengths in Illumina.
Informa UK Limited Online. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size. Typically Illumina paired end reads have an insert longer than the combined length of both reads see Figure Figure1 1.
However the average size of human coding exons is only 160bp. Here we modify the Illumina RNA ligation protocol to allow SS-PE sequencing by using a custom pre-adenylated 3 adaptor. 1Create reference transcriptome index for BowTie or BWA.
The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece. For instance for a PE150 sequencing run the insert size should be above 300bp. The sequencing starts at Read 1 Adapter mate 1 and ends with the sequencing from Read 2 Adapter mate 2.
Normally the insert size is longer than the sum of the two read lengths meaning there is an unsequenced inner part in the middle of the insert. The variation of insert sizes is often large and the average size difficult to control. This can result in a proportion of fragments with an insert size of less than the length of a single read.
RNA Biology Volume 9 Issue 5 Pages 596-609 Publisher. PE reads generally have a smaller insert size 1kp than MP 2-5 kb. -I 0 -X 500 BWA.
Insert sizes and read lengths 35300 bp allowing high resolution characterization of any genome. What is the optimal insert size. Paired end sequencing refers to the fact that the fragments sequenced were sequenced from both ends and not just the one as was true for first generation sequencing.
It depends on the used Illumina system the used reagents kits and the mode singlepaired end. Here we modify the Illumina RNA ligation protocol to allow SS-PE sequencing by using a custom pre-adenylated 3 adaptor. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size.
Selecting The Insert Size Of The Illumina Rna Seq For Paired-End Data Would Be. Standard paired-end libraries 200500 bp can be used to detect large and small insertions deletions indels inversions and other rearrangements. The Insert Length Should Fit At The End.
Illumina Systems have different DNA insert sizes for various tests ranging from 200 to 800 bpm. 2Align paired-end RNA-Seq data to the reference transcripts using minimum insert-length as zero and maximum insert-length as 500 or more. Insert size -- The insert size refers to the distance between the pairs.
For Illumina systems DNA insert size range of 200800 bp.
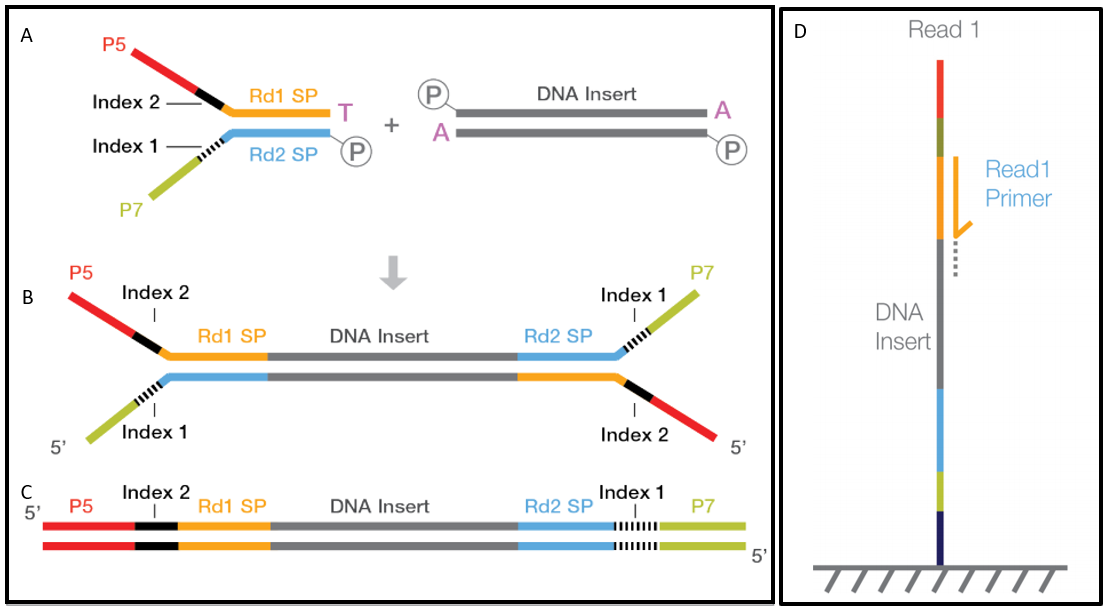
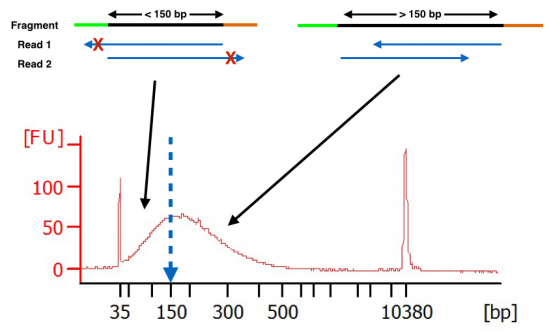
How Short Inserts Affect Sequencing Performance
How Short Inserts Affect Sequencing Performance
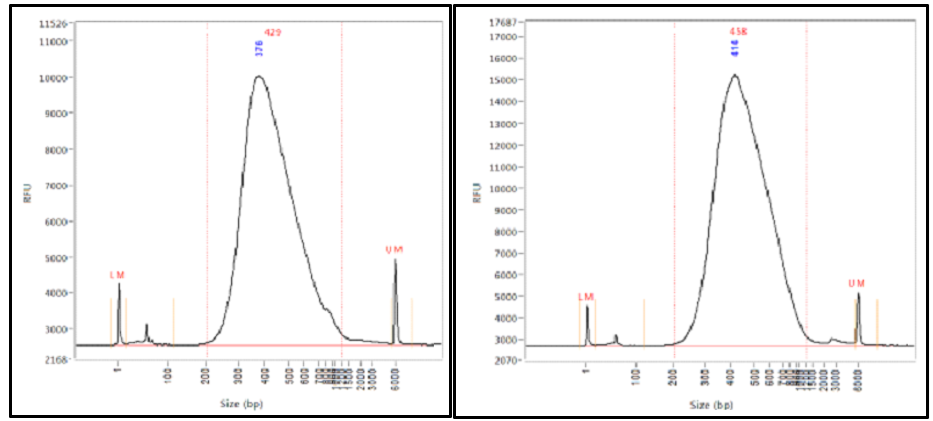
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

Interpreting Color By Insert Size Integrative Genomics Viewer

Five Approaches To Detect Cnvs From Ngs Short Reads A Paired End Download Scientific Diagram

Pdf Assessment Of Insert Sizes And Adapter Content In Fastq Data From Nexteraxt Libraries
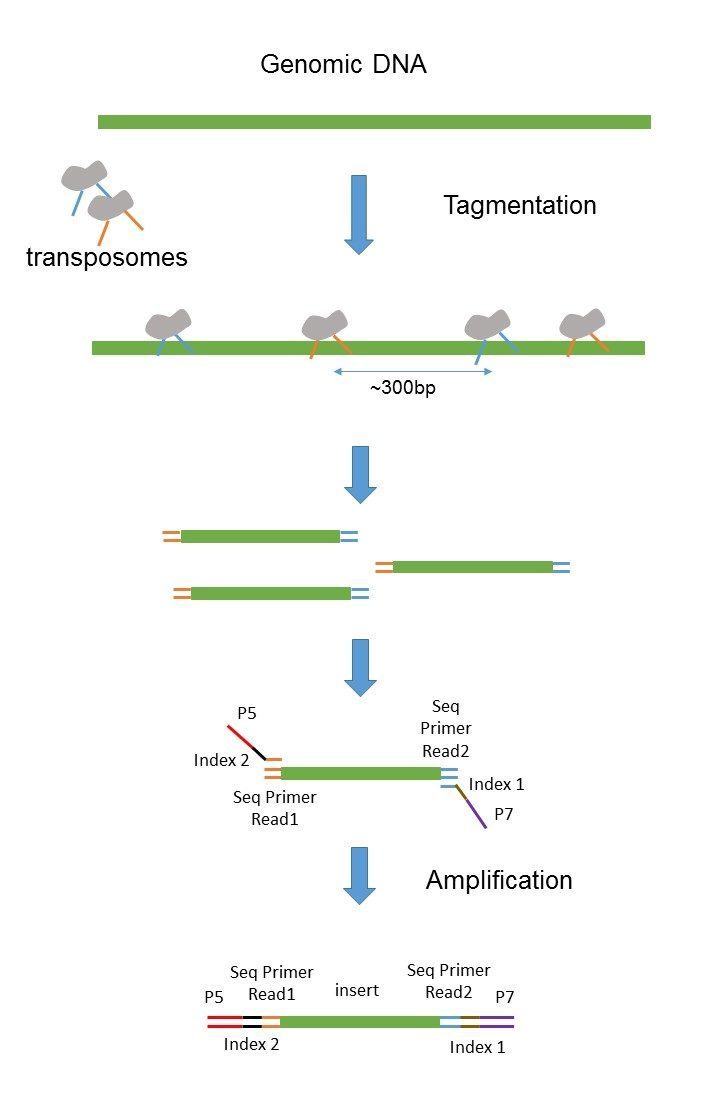
Paired End Sequencing France Genomique

Detecting Deletion Events A When Mapping Paired End Reads To The Download Scientific Diagram

What Are Paired End Reads The Sequencing Center

What Is Mate Pair Sequencing For

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

Library Insert Size Libraries Were Constructed Using The Standard A Download Scientific Diagram

Definition Of Essential Terms A Definition Of Fragment Read And Download Scientific Diagram
The Insert Size In Paired End Data Seqanswers

Interpreting Color By Insert Size Integrative Genomics Viewer
What Read Lengths Are Produced By Modern Illumina Sequencers